Examples¶
All sample scripts can be found in the folder timagetk/timagetk/examples/ and can be launched.
For example, to run the script input_output.py, open a shell prompt, go the
the folder examples and type: python input_output.py
Input/Output¶
This example illustrates input and output operations (see Package Components).
The resulting images (see folder timagetk/timagetk/examples/results/) can be checked against the reference image by using any image visualization tool (such as TissueLab or fiji).
# imports
try:
from timagetk.components import SpatialImage
from timagetk.components import imread, imsave
from timagetk.util import data_path
except ImportError:
raise ImportError('Import Error')
out_path = './results/' # to save results
# input image path
img_path = data_path('time_0_cut.inr')
# .inr format to SpatialImage
sp_img = imread(img_path)
# sp_img is now a SpatialImage instance
if isinstance(sp_img, SpatialImage):
# get the SpatialImage metadata, accessor
metadata = sp_img.get_metadata() # dictionary
# print the SpatialImage metadata
print('Metadata :'), metadata
# metadata modification
metadata['filename'] = img_path
# set the SpatialImage metadata, mutator
sp_img.set_metadata(metadata)
# print the SpatialImage metadata
print('New metadata :'), sp_img.get_metadata()
# get the SpatialImage voxelsize (list of floating numbers)
vox = sp_img.get_voxelsize()
# print the SpatialImage voxelsize
print('Voxelsize :'), vox
# voxelsize modification
new_vox = [] # empty list
for ind, val in enumerate(vox):
new_vox.append(2.0*val)
# set the SpatialImage voxelsize, mutator
sp_img.set_voxelsize(new_vox)
print('New voxelsize :'), sp_img.get_voxelsize()
# output
# filename
res_name = 'example_input_output.tif'
# SpatialImage to .tif format
imsave(out_path+res_name, sp_img)
# filename
res_name = 'example_input_output.inr'
# SpatialImage to .inr format
imsave(out_path+res_name, sp_img)
You can find a detailed description of:
- data structure
SpatialImage, see Module Spatial Image - functions
imreadandimsave, see Module Input/Output
Rigid, affine and deformable registration¶
This example illustrates rigid, affine and deformable registration (see Module Registration).
A registration process requires two input images (floating_img and reference_img) and computes:
- a spatial transformation
trsf_outbetween the two input images, - a result image
img_rescorresponding to the floating image resampled into the reference frame.
The resulting images (see folder timagetk/timagetk/examples/results/) can be checked against the reference image by using any image visualization tool (such as TissueLab or fiji).

# imports
try:
from timagetk.util import data_path
from timagetk.components import imread, imsave
from timagetk.plugins import registration
#from timagetk.wrapping.bal_trsf import BalTransformation
except ImportError:
raise ImportError('Import Error')
out_path = './results/' # to save results
# we consider two different times
# time_1 is the floating image
# time_2 is the reference image
times = [1, 2]
# list of SpatialImage instances
list_images = [imread(data_path('time_' + str(time) + '.inr'))
for time in times]
floating_img, reference_img = list_images[0], list_images[1]
# Rigid registration:
trsf_rig, res_rig = registration(floating_img,
reference_img,
method='rigid_registration')
# display the spatial transformation (4x4 matrix):
trsf_rig.c_display()
# save the spatial transformation:
res_name = 'example_trsf_rigid.trsf' # filename
trsf_rig.write(out_path+res_name)
# save the result image:
res_name = 'example_reg_rigid_1_2.tif' # filename
# SpatialImage to .tif format
imsave(out_path+res_name, res_rig)
# Affine registration:
trsf_aff, res_aff = registration(floating_img,
reference_img,
method='affine_registration')
res_name = 'example_reg_affine_1_2.tif' # filename
# SpatialImage to .tif format
imsave(out_path+res_name, res_aff)
# Deformable registration:
trsf_def, res_def = registration(floating_img,
reference_img,
method=
'deformable_registration')
res_name = 'example_reg_deformable_1_2.tif' # filename
# SpatialImage to .tif format
imsave(out_path+res_name, res_def)
# Save the reference image:
res_name = 'example_reg_reference.tif' # filename
# SpatialImage to .tif format
imsave(out_path+res_name, reference_img)
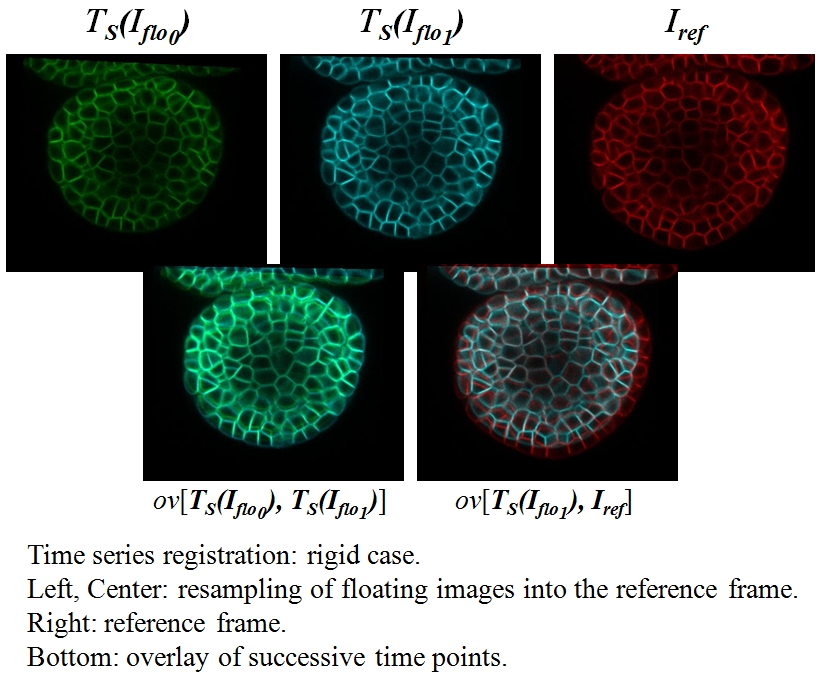
Sequence Registration¶
This example illustrates rigid and affine registration along a whole sequence (see Module Sequence registration).
This process requires a list of images (a sequence) and computes:
- a list of spatial transformations
list_compo_trsfbetween successive images and the reference image, - a list of result images
list_res_imgcorresponding to the successive images resampled into the reference frame.
By default, the reference image is the last image of the sequence and all images are resampled into this frame. The resulting images (see folder timagetk/timagetk/examples/results/) can be checked against the reference image by using any image visualization tool (such as TissueLab or fiji).
# imports
try:
from timagetk.util import data_path
from timagetk.components import imread, imsave
from timagetk.plugins import sequence_registration
except ImportError:
raise ImportError('Import Error')
out_path = './results/' # to save results
# we consider three different times
# time_0 is the floating image
# time_1 is first the local reference image,
# and becames the floating image
# time_2 is the global reference image
times = [0, 1, 2]
# list of SpatialImage instances
list_images = [imread(data_path('time_' + str(time) + '.inr'))
for time in times]
# Rigid registration along the whole sequence:
# list_compo_trsf : list of transformations
# list_res_img : list of resulting images
# the primitive embeds an iterative registration
list_compo_trsf, list_res_img = sequence_registration(list_images,
method='sequence_rigid_registration')
for ind, img in enumerate(list_res_img):
# filenames
res_name = ''.join(['example_seq_reg_rigid_', str(ind), '_',
str(times[-1]), '.inr'])
# SpatialImage to .inr format
imsave(out_path+res_name, img)
# Affine registration alog the whole sequence:
list_compo_trsf, list_res_img = sequence_registration(list_images,
method='sequence_affine_registration')
for ind, img in enumerate(list_res_img):
# filenames
res_name = ''.join(['example_seq_reg_affine_', str(ind), '_',
str(times[-1]), '.inr'])
# SpatialImage to .inr format
imsave(out_path+res_name, img)
Seeded-watershed segmentation¶
This example illustrates seeded-watershed segmentation.
# imports
try:
from timagetk.util import data_path
from timagetk.components import imread, imsave
from timagetk.plugins import linear_filtering, morphology
from timagetk.plugins import h_transform
from timagetk.plugins import region_labeling, segmentation
from timagetk.plugins import labels_post_processing
except ImportError:
raise ImportError('Import Error')
out_path = './results/' # to save results
# we consider an input image
# SpatialImage instance
input_img = imread(data_path('input.tif'))
# optional denoising block
smooth_img = linear_filtering(input_img, std_dev=2.0,
method='gaussian_smoothing')
asf_img = morphology(smooth_img, max_radius=3,
method='co_alternate_sequential_filter')
ext_img = h_transform(asf_img, h=150,
method='h_transform_min')
con_img = region_labeling(ext_img, low_threshold=1,
high_threshold=150,
method='connected_components')
seg_img = segmentation(smooth_img, con_img, control='first',
method='seeded_watershed')
# optional post processig block
pp_img = labels_post_processing(seg_img, radius=1,
iterations=1,
method='labels_erosion')
res_name = 'example_segmentation.tif'
imsave(out_path+res_name, pp_img)
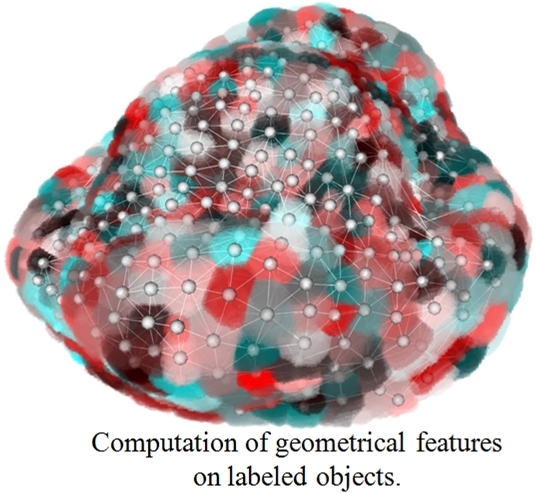
Computation of features¶
This example illustrates computation of geometrical features on labeled images. This process requires a labeled image and computes several features such as moments, spatial relationships, etc. This algorithm returns a dictionary that can be pickled (see pickle module).
# imports
import numpy as np
try:
from timagetk.util import data_path
from timagetk.components import imread
from timagetk.algorithms import GeometricalFeatures
except ImportError:
raise ImportError('Import Error')
try:
import cPickle as pickle
except:
import pickle
out_path = './results/' # to save results
# We consider two different times
times = [3,4]
list_images = [imread(data_path('time_' + str(time) + '_seg.inr'))
for time in times] # list of SpatialImage
feature_space_list = []
for ind, time in enumerate(times):
segmentation = list_images[ind] # segmentations
labels = np.unique(segmentation).tolist() # labels
background_id = np.min(labels)
labels.remove(background_id) # remove background
int_seg = GeometricalFeatures(segmentation, label=labels) # GeometricalFeatures object
feature_space_dict = int_seg.compute_feature_space(background_id=background_id) # compute feature space
feature_space_list.append(feature_space_dict)
for ind, time in enumerate(times):
pkl_path = ''.join([out_path, 'example_geom_feat_time_', str(times[ind]), '.pkl'])
out = open(str(pkl_path), 'wb')
# save as pickle object (serialization)
pickle.dump(feature_space_list[ind], out)
out.close()
print('')
sp_label = 137
print('Retrieve information for the label :'), sp_label
print('')
print('Label : '), str(sp_label), feature_space_list[0][sp_label]
print('')
print('Label : '), str(sp_label), feature_space_list[1][sp_label]
Label : 137 {'Neighbors': [32, 124, 149, 195, 282, 297, 298, 312],
'Distances': {32: 5.28, 195: 6.963, 297: 4.348, 298: 5.989,
149: 5.8, 312: 4.783, 282: 5.665, 124: 7.703},
'Bounding box volume': 12180,
'Physical centroid': (55.428, 62.878, 1.191),
'Label': 137, 'Bounding box size': (29, 30, 14),
'Physical volume': 117.329, 'Index centroid': (186, 211, 3),
'Bounding box': (172, 200, 197, 226, 0, 13)}
Temporal tracking¶
This example illustrates tracking of objects on labeled images.
# imports
import numpy as np
try:
from timagetk.util import data_path
from timagetk.components import imread, imsave, SpatialImage
from timagetk.plugins import labels_post_processing
from timagetk.algorithms import GeometricalFeatures
from timagetk.algorithms import TemporalMatching
except ImportError:
raise ImportError('Import Error')
try:
import cPickle as pickle
except:
import pickle
out_path = './results/'
# We consider two different times
times = [3,4]
segmentation_list, feature_space_list, back_id_list = [], [], []
for ind, val in enumerate(times):
img = imread(data_path('time_' + str(val) + '_seg.inr'))
# subimage extraction
shape = img.get_shape()
indices = [0,shape[0]-1,0,shape[1]-1,0,5]
img = img.get_region(indices=indices)
# remove small cells
labels = np.unique(img).tolist()
img = labels_post_processing(img, method='labels_erosion', radius=2)
img[img==0] = np.min(labels)
img = SpatialImage(img, voxelsize=img.get_voxelsize())
# save input labeled images
res_name = 'example_track_time_' + str(val) + '_seg.inr'
imsave(out_path+res_name, img)
labels = np.unique(img).tolist() # list of labels
back_id = np.min(labels) # background identifier
back_id_list.append(back_id)
labels.remove(back_id)
# feature space computation
obj_gf = GeometricalFeatures(img, label=labels)
feature_space = obj_gf.compute_feature_space()
segmentation_list.append(img), feature_space_list.append(feature_space)
# temporal matching
obj_tm = TemporalMatching(segmentation_list=segmentation_list,
feature_space_list=feature_space_list,
background_id_list=back_id_list)
# computation of admissible matching
adm_match_list = obj_tm.get_admissible_matching_list()
# matching criterion (default is None)
criterion='Jaccard coefficient'
# computation of normalized matching costs
norm_cost_list = obj_tm.compute_normalized_cost(adm_match_list,
criterion=criterion)
poss_opt = ['matching','iterative_matching']
opt = 'matching'
if opt in poss_opt:
if opt=='matching':
# appear/disappear cost
alpha_cost = int((80/100.0)*obj_tm.max_cost)
# matching
matching_dict = obj_tm.matching(norm_cost_list, alpha_cost)
elif opt=='iterative_matching':
# initial graph
init_graph = obj_tm.build_init_graph(norm_cost_list)
# iterative matching
matching_dict = obj_tm.iterative_matching(norm_cost_list, init_graph)
# write a dictionary of matching
pkl_path = ''.join([out_path, 'example_track_matching_dict.pkl'])
pkl_file = open(str(pkl_path), 'wb')
# save as pickle object (serialization)
pickle.dump(matching_dict, pkl_file)
pkl_file.close()
# resulting images
img_list = obj_tm.res_images(matching_dict)
# save results
res_name = 'example_track_initial_cells.inr'
imsave(out_path+res_name, img_list[0])
res_name = 'example_track_final_cells.inr'
imsave(out_path+res_name, img_list[1])
res_name = 'example_track_disapp_cells.inr'
imsave(out_path+res_name, img_list[2])
res_name = 'example_track_app_cells.inr'
imsave(out_path+res_name, img_list[3])
else:
print('Poss. opt :'), poss_opt
